Molecular Biological Detection of Pathogens
FISH – Fluorescence in situ Hybridization
FISH – Fluorescence in situ Hybridization
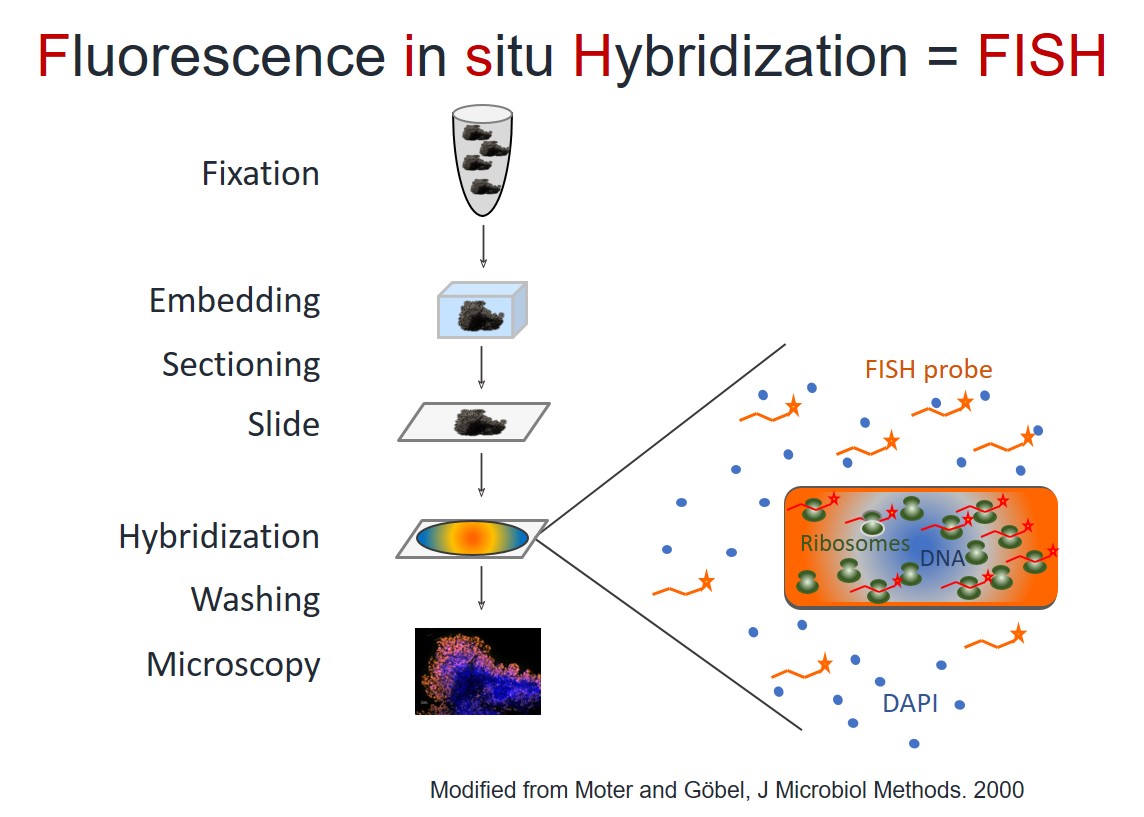
FISH is a microscopic method, which combines the advantages of molecular biology, fluorescence microscopy and histology.
FISH allows imaging of microorganisms in the sample tissue context. In addition, FISH identifies the microorganisms and quantifies their amount, localization and activity.
FISH is based on fluorescently labeled probes that bind sequence specific to the ribosomal RNA (rRNA) of the microorganisms. Thus, FISH visualizes bacteria and fungi and detects them on genus- or species levels. For example, probes may detect all bacteria, all staphylococci or just Staphylococcus aureus. FISH measures the (remaining) activity of the microorganisms based on their ribosome content on a single cell-level. Successful treatment of microbial biofilms with antimicrobial substances decreases the fluorescence signal when compared to active biofilms. Therefore, FISH assesses the efficacy of antimicrobial substances by measuring the reduction of the FISH-positive fraction as compared to the entire biofilm mass. We quantify the effect by digital image analysis.
FISH is therefore an ideal method for the analysis of biofilm, the testing of antimicrobial substances against biofilms and the development of innovative anti-biofilm materials.
Which information does FISH give?
- Identification
- Localization
- Organisation
- Activity of microorganisms
Which advantages does FISH have?
- Identification of pathogens in culture-negative cases
- Differentiation between contamination and infection
- Amount and localization of pathogens
- Organization of microorganisms (planktonic or biofilm?)
- Identification of key pathogen(s) in multi-species infections
- Measurement of microbial activity
- Effectiveness measurement of antimicrobial substances or procedures
Molecular Amplification Techniques
PCR | Sequencing | NGS | Microbiome analysis | 16S rRNA gene
16S rRNA gene PCR amplification and Sanger sequencing
We offer classical pan-bacterial amplification of the 16S rRNA gene with subsequent Sanger sequencing for the identification of bacteria from culture or tissue samples.
PCR (polymerase chain reaction) amplifies parts of the genetic material of microorganisms (for example part of the 16S rRNA gene). The amplicons are then sequenced (either by Sanger sequencing or through microbiome analysis), so that the microorganisms can be identified.
Next generation sequencing (NGS) and microbiome analysis
Microbiome analysis investigates all microorganisms present in a given sample. Before bioinformatics analysis of the amplification results, several steps have to take place: DNA extraction, PCR amplification, Bar-coding and Next Generation Sequencing. We offer microbiome analysis for mixed cultures or multi-species samples. This method is particularly useful for mixed biofilms.
FISH-Seq
We detect microorganisms with a so far unmatched precision and sensitivity by combining FISH with PCR amplification and sequencing out of methacrylate sections from the same sample.
FISHseq offers the following advantages:
- Unmatched reliability of results by correlation of the microscopy with sequencing data
- Unmatched sensitivity through mechanical sectioning before DNA extraction
- Unmatched specificity through avoidance of contamination of microorganisms and DNA during sample processing
