Molecular Biological Services and Biofilm Research
We analyze medical biofilms by Fluorescence in situ Hybridization (FISH) and nucleic acid amplification techniques (PCR, sequencing, microbiome analysis). The combination of imaging with genome/microbiome analysis provides innovative insights into infection landscapes and biofilms. For our diagnostic spectrum, please see: www.moter-diagnostics.com/en/diagnostic-spectrum
We analyze samples ranging from in vitro experiments to tissue samples. We focus on the quantification and analysis of biofilms. Here, the FISH method shows the efficacy of anti-biofilm substances or coated surfaces of medical products (for example implants) against microorganisms.
Visualization and Quantification of Biofilms
Assessment of microbial biofilms
Fluorescence in situ Hybridization (FISH) visualizes biofilms microscopically and assesses their extent, thickness and localization.
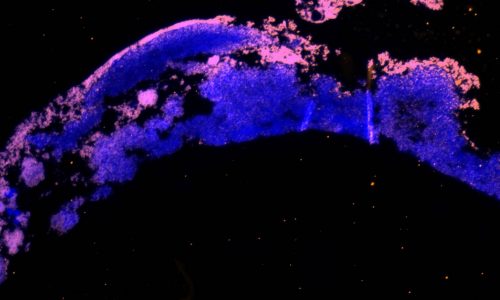
FISH of a bacterial biofilm (blue – nucleic acid stain DAPI showing all bacteria, magenta – FISH-positive bacteria).
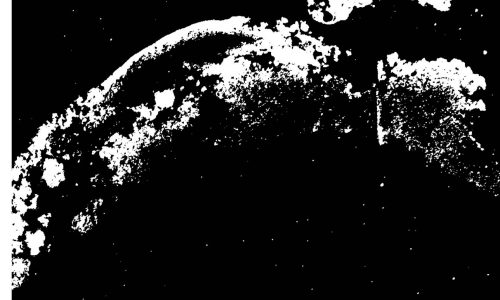
Image segmentation by digital image analysis for the quantification of the FISH-positive biofilm fraction.
Standardized FISH Tests
for Antimicrobial Substances
Fluorescence in situ Hybridization for the efficacy measurement of antimicrobial substances
Fluorescence in situ Hybridization (FISH) is a molecular biological method, which allows measuring the efficacy of antimicrobial substances against biofilms by imaging and visualization.
FISH also is suitable for testing anti-infective substances. Thus, FISH quantifies the antimicrobial’s effect culture-independent and with a spatial dimension: where is the substance effective? How does the substance work? Do surviving microorganisms remain? How dependent on time and dose is the antimicrobial’s effect in a biofilm?
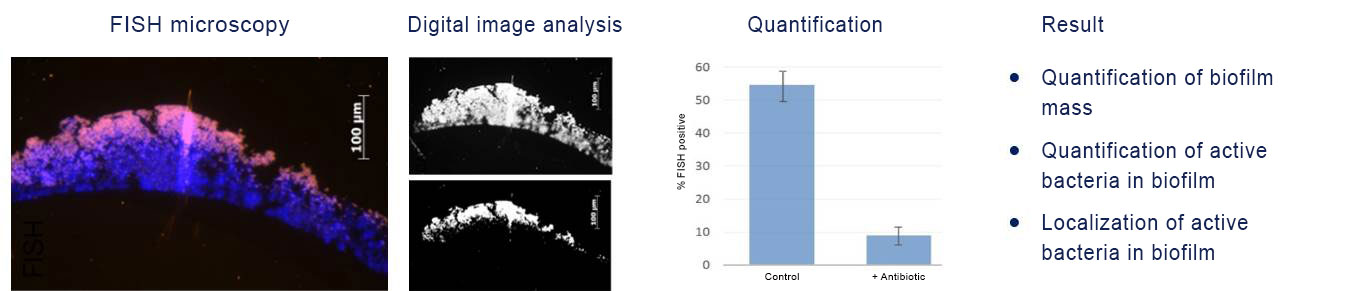
FISH of staphylococcal biofilm. Detection of the in situ efficacy of antibiotics on biofilms (DAPI, blue). The fraction of FISH-positive bacteria (magenta) is reduced upon antibiotic treatment as compared to the control.
Testing the Antimicrobial Properties of
Medical Surfaces
Visualization and quantification of biofilms by FISH
Fluorescence in situ Hybridization (FISH) visualizes and measures the effect of antimicrobial-coated surfaces against microorganisms and biofilms. FISH quantifies the antimicrobial’s effect culture-independent and with a spatial dimension, for example on coated surfaces of medical implant.

Catheter cross-sections with attached biofilms, embedded in methacrylate.
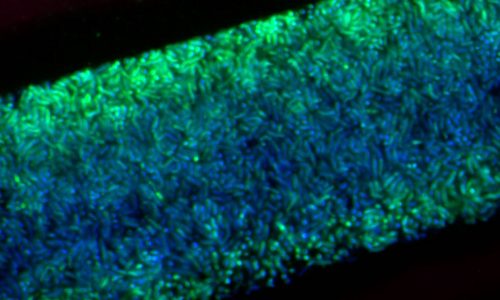
Surface-attached microbial biofilm with layers of differently active microorganisms. Active bacteria show a strong FISH-signal (green), whereas inactive bacteria only show the blue signal of the nucleic acid DAPI.
FISH Analysis of Microorganisms – Custom FISH
Development of new, individual FISH probes and assays
For research projects, we individually develop customized FISH assay solutions. The applications range from flow-chamber systems to tissue samples.
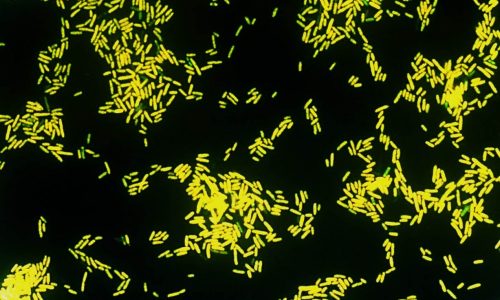
FISH of Escherichia coli (Gram-negative rod).
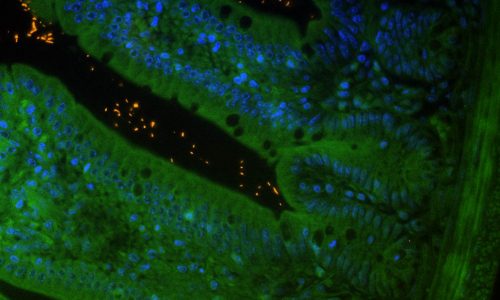
FISH analysis of intestinal microorganisms in the lumen of a murine gut cross-section. Tissue background in green (autofluorescence), nucleic acids in blue (nucleic acid stain DAPI), FISH-positive bacteria in orange (EUB338 probe).
Molecular Identification of Microorganisms
16S rRNA gene PCR amplification and Sanger sequencing
For the identification of microorganisms from culture or out of tissue samples, we offer classical pan-bacterial PCR amplification of the 16S rRNA gene followed by Sanger sequencing. PCR (polymerase chain reaction) amplifies parts of the genetic material of microorganisms (for example part of the 16S rRNA gene), for which subsequently sequence analysis reveals the identity of the microorganism.
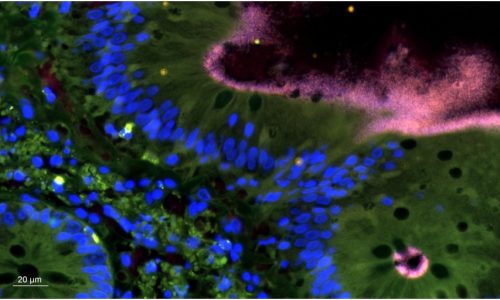
FISH reveals the presence of Brachyspira sp. in a tissue sample.
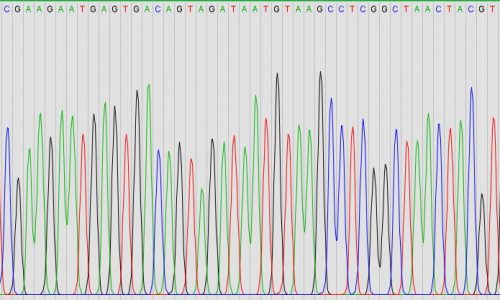
MG-FISH (Microbiome-guided FISH)
Microbiome-guided (MG) FISH
In mixed biofilms, we combine FISH with microbiome analysis, which identifies all microorganisms in a given sample (the entire microbiome).
We amplify microbial DNA out of methacrylate-embedded samples with a very high sensitivity and without contamination. Therefore, we interpret FISH and sequencing results together, which allows for a very high technical and diagnostic accuracy.
FISH gives the microbiome data a spatial dimension in the context of the intact sample:
- Quantity and distribution of the microorganisms involved
- Localization at and in the tissue
- Identification of key pathogens
- Spatial resolution of the sample (first colonizers on implants? Tissue-invasive pathogens?)
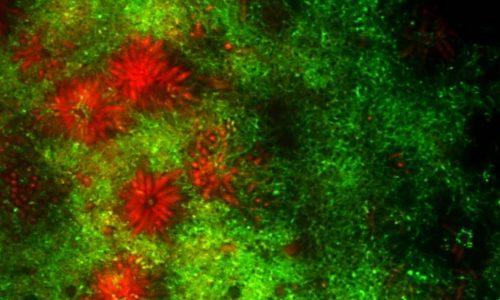
FISH of an oral multi-species biofilm.
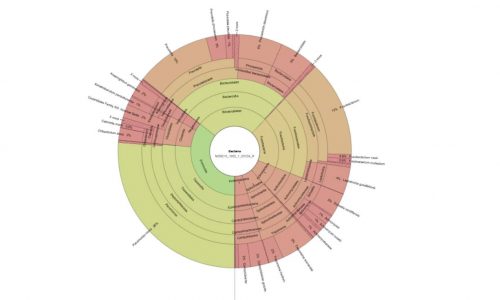
Microbiome analysis out of sections of the oral biofilm.
